Promotional Features
Molecular detection of beer-spoilage bacteria
Beer-spoilage microorganisms cause an increase of turbidity and unpleasant flavors in beer. Since improved process technology in modern breweries significantly reduced oxygen content in the final product, the role of strictly anaerobic bacteria like Pectinatus and Megasphaera has become more prevalent.
Detection of these organisms, which is traditionally carried out by incubation on culture medium, can take a week or more. A rapid molecular test system is therefore desirable for the detection of all known beer-spoiling microorganisms in one single test. Results of this study on a rapid test kit demonstrate that all beer-spoiling microorganisms can be detected and identified in a shorter time and in one test only.
Beer quality
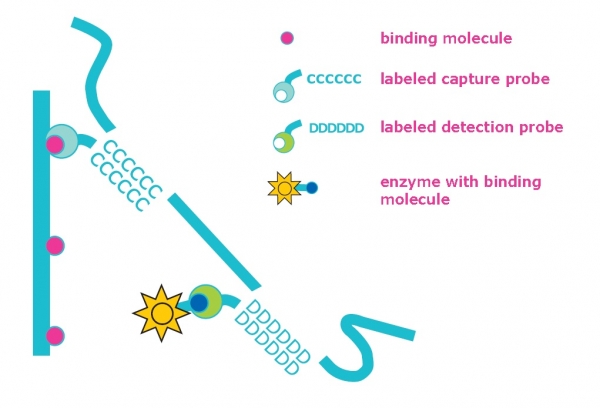
It is important to maintain quality and enhance the stability of beer in the production process through early detection of beer-spoiling microbes. Known beer-spoiling species and genera such as Lactobacillus, Pediococcus, Pectinatus and Megasphera can all be detected using the HybriScan D Beer rapid molecular test system in one single test. This beer test is based on the detection of target molecules from the microorganisms of interest by means of specific capture and detection probes using sandwich hybridization. The hybridization reaction of the target molecules with the Biotin-labeled capture and a digoxigenin-labeled detection probe takes place in a streptavidin-coated microtiter plate (see Figure).
After coupling of the target molecule to the microtiter plate, an enzyme is attached in a subsequent incubation step. After several washing steps, reaction with a color substrate gives blue coloration that changes into yellow color after the addition of a stop solution. The yellow color enables optional highly sensitive photometric measurement at 450 nm. Qualitative results can be determined by eye or, for quantitative measurement, use with a microplate reader. Standard solutions are provided as controls in the test kit.
Experimental method to analyze beer samples for spoilage organisms
In this study, beer samples with alcohol content between 2.5% and 6.7% were tested with the HybriScan D beer test kit. The beer samples were spiked with L. brevis, L. coryniformis, L. lindneri, Ped. damnosus, M. cerevisiae and Pec. frisingensis in a range between 103 and 106 CFU/sample. The bacteria were cultivated in media according to DSMZ guidelines. The cultures were harvested and washed with sterile saline solution. Bacterial counts were then determined by measuring OD600 and by counting colony forming units (CFU) on agar. Beer samples were then inoculated with the bacterial cultures.
Thirty additional brewery samples were also examined using the HybriScan D beer test for beer-spoiling microorganisms. Up to 10 mL of the beer sample was processed according to the HybriScan D protocol. The results were controlled by cultivation on NBB agar.
For the development of further genera and species, specific test solution experiments were performed with 11 additional pure bacterial cultures of most common beer-spoiling micro-organisms for detection by HybriScan D test experiments. These bacterial strains were also cultivated on media according to DSMZ protocols and processed according to HybriScan D protocol.
Results
In this study, a total of 178 samples, with alcohol content between 2.5% and 6.7%, were tested with HybriScan D beer test. In order to confirm that it is possible to detect different beer-spoiling species with the beer test solution, the samples were spiked with L. brevis, L. coryniformis, Ped. damnosus, Pec. frisingensis and M. cerevisiae. Results were compared with the inoculated quantity of bacteria. The bacterial counts in the beer samples determined by the HybriScan D test were in the same range as the inoculated values. Pec. frisingensis was only detected in beer samples with 2.5% alcohol.
Results of the HybriScanD test were below the counts of the inoculum. The number of microbes in beer is possibly diminished because the strain used in the study was not adapted to beer.
Pec. frisingensis were not detected in beer with a higher alcohol content (data not shown). It was observed that beer with a low alcohol content is more prone to spoilage with Pectinatus and Megasphaera species.1 Pectinatus species are more resistant and can survive in acidic conditions. The pH tolerance of these anaerobic bacteria is influenced by the presence of ethanol.2 Real beer samples from the brewery were also tested with the HybriScan D beer test for beer-contaminating bacteria. These samples were also simultaneously checked with NBB agar. The HybriScan D test gave a positive signal for each sample. These results were confirmed on agar.
For a more sophisticated analysis, seven additional tests to specifically test for the following organisms: “Pectinatus spp./Megasphaera spp.”, “Pediococcus spp.”, “Lactobacillus brevis”, “L. lindneri”, “L. collinoides”, “L. rossiae” and “L. backii“ were developed and optimized. These optimizing experiments were performed with 17 beer-spoiling bacteria strains. A microtiter plate with eight different test solutions were tested with a positive and negative control and 10 beer-spoiling bacteria species. The experiment was performed with bacterial cultures which contained bacterial amounts ≥106 cfu/sample.
The beer test solution gave a positive signal with all used bacteria. Yellow signals in the microtiter plate confirmed that the optimized test solutions react in a species-specific manner and showed no cross-reactivity. The experiments for the optimization of the test solutions indicated that the detection limit of those test solutions differs for Lactobacillus spp. And the other mentioned beer spoiling genera between 104 and 105 CFU/sample.
Conclusions
This study confirms the following main conclusions about the performance of the HybriScan D Beer test:
- It is a rapid and sensitive molecular biological test system that is comparable to the classical cultivation method.
- It offers qualitative and quantitative detection of the most well-known beer-spoiling organisms in one single test.
- It is flexible as it allows combination with different read-out technologies, and detection of rRNA and mRNA is possible.
- It is a robust test with easily predictable specificity of the probes, and easy synthesis of the oligonucleotide probes.
- It is a fast and cost-effective test with inexpensive read out device technology.
References
1. A. D. Paradh, W. J. Mitchell and A. E. Hill J. Inst. Brew. 117(4), 498 - 506 (2011).
2. Suzuki, K., 125th Anniversary review: Microbiological instability of beer caused by spoilage bacteria. J. Inst. Brew., 2011, 117, 131-155 (2011).
HybriScan is a registered trademark of ScanBec GmbH.